Background
About six months ago, I wrote an article about the SARS-CoV-2 Genome Assembly. In this article, I published a script that aimed to reproduce the findings by Wu et al., 2020, which lead to the first ever published genomic sequence of the SARS-CoV-2 virus, upon which the whole pandemic is based on.
My two main findings back then were:
There are three posted versions of the genome, and none of these were exactly reproducible by running Megahit (the de-novo assembler used to produce the longest contig used for the genome).
The full genome was not reproducible by Megahit alone. Even the authors never claimed that, as they said they used RACE to determine “the complete genome termini”.
For context: “RACE can provide the sequence of an RNA transcript from a small known sequence within the transcript to the 5' end (5' RACE-PCR) or 3' end (3' RACE-PCR) of the RNA.” Essentially, a small region of the end of the contig is used for a PCR primer, to allegedly find the end of the genome. How it is possible, to know where really the end is, and if the assembler has really stopped the assembly close to the end, remains unclear.
While I succeeded in assembling the core of the genome, the produced contigs differed significantly in length from what Wu et al. had published. In the comments of my article, some people pointed out that I was missing a step in my pipeline: “Sequencing reads were first adaptor and quality trimmed using the Trimmomatic program”.
Besides many of the open questions that I have raised in my previous article, the few main issues with the Wu et al. 2020 paper are:
Not able to reproduce the 384,096 contigs based on the source data, (only 22,566 contigs).
There are three versions of the claimed genome uploaded to Genbank
The longest contig is claimed to have: “30,474 nucleotides (nt)”, but the three versions of the MN908947 only list a contig with 30,473 base pairs.
Overview of my Seven Part Genomics Series:
Methods & Results
As the quality trimming step was missing earlier, I have now updated the script, and re-ran the assembly. Here are the results, along with some more information and understanding I have gathered along the way.
The additional trimming step yielded this output:
[ec2-user@ip-172-31-17-69 ~]$ java -jar Trimmomatic-0.39/trimmomatic-0.39.jar PE SRR10971381_{1,2}.fastq {1,2}{,un}paired.fastq.gz AVGQUAL:20 HEADCROP:12 LEADING:3 TRAILING:3 MINLEN:75 -threads 32
TrimmomaticPE: Started with arguments:
SRR10971381_1.fastq SRR10971381_2.fastq 1paired.fastq.gz 1unpaired.fastq.gz 2paired.fastq.gz 2unpaired.fastq.gz AVGQUAL:20 HEADCROP:12 LEADING:3 TRAILING:3 MINLEN:75 -threads 32
Quality encoding detected as phred33
Input Read Pairs: 28282964 Both Surviving: 13785883 (48.74%) Forward Only Surviving: 96245 (0.34%) Reverse Only Surviving: 27364 (0.10%) Dropped: 14373472 (50.82%)
TrimmomaticPE: Completed successfullySo now, that we have “quality trimmed” the reads, let’s run Megahit again. As noted in the earlier article, Wu ran Megahit with version 1.1.3, while the latest version even at that time was already 1.2.9 - hence following are the results for both versions.
Megahit 1.2.9
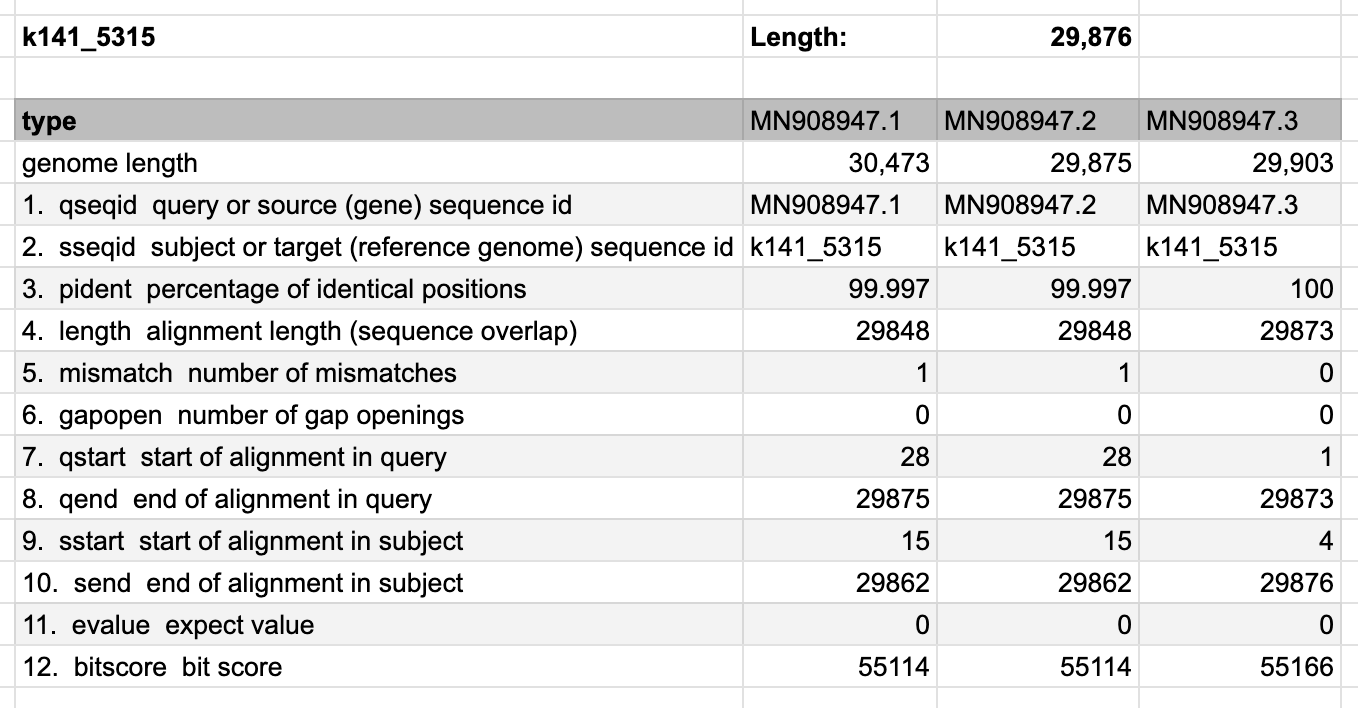
With the current version of Megahit, it assembles the longest contig with 29,875 base pairs.
2023-03-26 17:23:14 - 22169 contigs, total 11114272 bp, min 200 bp, max 29875 bp, avg 501 bp, N50 473 bpHowever, comparing this contig to the three versions of Wu’s genome, we don’t get a perfect match either. The first version has many more base pairs and there is one mismatch. The second version fits the total length, but only 29,848 base pairs match, except a single mismatch. The third version is probably the closest, as there’s no more mismatch, hence 100% identical positions for 29,873 base pairs, but the total length is off.
Upon closer inspection of the difference to the third genome, we can see, that only the first two letters are different, and the tail is missing (which was likely manually added):
$ cat k141_7410.fasta MN908947.3.fasta|mafft -|awk '{printf(/^>/?"\n%s\n":"%s"),$0}'|grep .>temp.aln;paste <(sed -n 2p temp.aln|grep -o .) <(sed -n 4p temp.aln|grep -o .)|awk '$1!=$2{print NR,$1,$2}'
1 g -
2 g -
29876 - a
29877 - a
29878 - a
29879 - a
29880 - a
29881 - a
29882 - a
29883 - a
29884 - a
29885 - a
29886 - a
29887 - a
29888 - a
29889 - a
29890 - a
29891 - a
29892 - a
29893 - a
29894 - a
29895 - a
29896 - a
29897 - a
29898 - a
29899 - a
29900 - a
29901 - a
29902 - a
29903 - a
29904 - a
29905 - aMegahit 1.1.3
The version that Wu et al. documented to have used, yielded this result, which did not even come close either to the originally claimed longest contig of 30,474 bp:
--- [STAT] 22566 contigs, total 11041148 bp, min 200 bp, max 29876 bp, avg 489 bp, N50 463 bpComparing this contig to the three versions of Wu’s genome again. The first version has many more base pairs, there’s one mismatch. The second version totals one base pair fewer, but only 29,848 base pairs match, including 1 mismatch. The third version against is the closest, as there’s no more mismatch, hence 100% identical positions for 29,873 base pairs, but the total length is off again.
Let’s also inspect the differences to the third version again, and we get a similar picture as before, except this time the first three base pairs are different, and the tail is missing again.
$ cat k141_5315.fasta MN908947.3.fasta|mafft -|awk '{printf(/^>/?"\n%s\n":"%s"),$0}'|grep .>temp.aln;paste <(sed -n 2p temp.aln|grep -o .) <(sed -n 4p temp.aln|grep -o .)|awk '$1!=$2{print NR,$1,$2}'
1 g -
2 g -
3 g -
29877 - a
29878 - a
29879 - a
29880 - a
29881 - a
29882 - a
29883 - a
29884 - a
29885 - a
29886 - a
29887 - a
29888 - a
29889 - a
29890 - a
29891 - a
29892 - a
29893 - a
29894 - a
29895 - a
29896 - a
29897 - a
29898 - a
29899 - a
29900 - a
29901 - a
29902 - a
29903 - a
29904 - a
29905 - a
29906 - aDiscussion
I have shown that, even with the previously missing step of quality trimming of the reads, I can still not reproduce the exact results, that Wu et al 2020 have documented in their paper. To this date, and as to my knowledge, no one has been able to reproduce the longest claimed 30,474 contig from the provided raw data.
Furthermore, a commenter in my last article mentioned, that Wu might have run the original assembly on the unfiltered reads (for a human genome match), which unfortunately are not published for full transparency. The commenter also mentioned, that even the filtered reads, after the latest update to the human genome, yield some more matches, that would have previously not been filtered out. While I have not personally tried to verify this yet - if true - this would be concerning, because we don’t know what future updates to the human genome would reveal, and if the reads are really of non-human origin.
While it is possible to reproduce almost the entire claimed genome with Megahit, it is still clear, that the tail is always missing, and other assemblers have failed to even come close to these results. This has also been confirmed as such by Islam, et al. 2021, which state, that “[…] the entire viral genome was not assembled in most cases, especially at the termini of the genome.”
They go on, to ask for better assemblers, and then state this circular fallacy: “The SARS-CoV-2 virus genome could also be assembled by aligning the reads to the reference genome or using a reference guided assembly.” Aligning reads, means, to align the input reads again to the assembled longest contig (or final genome, including the termini). Obviously this is a fallacy, as the reference genome (the one we discussed here, by Wu et al. 2020) is assembled by the exact same method (de-novo assembly, Megahit) that failed to assemble the entire genome in the first place.
Another interesting aspect is the question of isolation, that appears to very controversial, and has brought me many Twitter discussions. From the Wu et al. paper it becomes clear, for the vigilant reader, that Wu et al. clearly did not have a purified isolate, otherwise there would be no need to programmatically filter out reads that matched the human genome.
Open Questions
Lastly, in my last article I also raised many important questions, which were not answered sufficiently by anyone to this date, and which I have amended by a few additional ones:
Why was the claimed genome updated so many times (three times)?
Why did the length change?
Why does the paper mention 30,474 nt, when the longest deposited contig was only 30,473 nt?
Why was the patient moved between hospitals while being in ICU on a ventilator, and what was his final outcome?
Is it certain, that all that is left in the dataset is of non-human origin?
Why did Wu et al. use the much older version 1.1.3 of Megahit, instead of the latest 1.2.9 version?
Why did Trinity and Megahit produce so vastly different results?
Why can the longest original contig (30,474nt) not be assembled with Megahit?
Why is no other de-novo assembler (e.g. trinity, spades) even remotely able to assemble the full genome?
Why has the full/entire genome not been validated against the original patient sample (or other samples)?
Why was the longest contig chosen, what about the other contigs of the over 384 thousand assembled contigs?
Why does Megahit only assemble about 22 thousand contigs, when Wu et al claims 384 thousand?
How is it established, with certainty, that the contig is of viral origin?
Why are the unfiltered reads not published?
Why is Megahit not able to assemble the tail?
How can it be ruled out that no other undetected pathogen is responsible for the illness (of the other 1.7M contigs)?
How it is possible, to know where the actual end of the genome is and if the assembler has really stopped the assembly close to the end?
Summary
So in summary we can conclude, that neither the results of the Wu et al. 2020 paper, is precisely reproducible to this date, nor has the SARS-CoV-2 genome been completely assembled by a single method and/or assembler. In addition, there remain many relevant unanswered questions, that should be answered by the responsible persons in charge.
A big thanks to reader Mongol and Paul for their fruitful ongoing discussions.
More Info
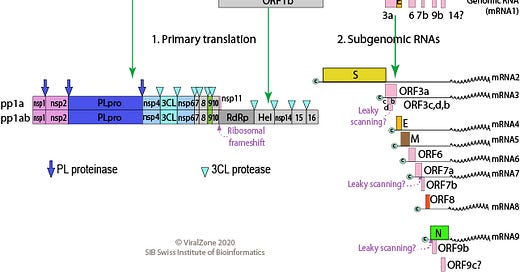
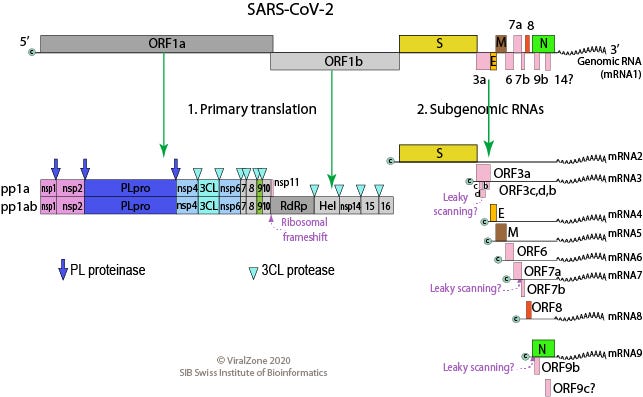
The Megahit assembler, mentioned in this article uses the “de-Bruijn graph” approach for de-novo assembly, to quickly understand how this works in practice, check out this great short video:
Sources & Code
You can find all code and results mentioned in this article here:
Further resources:
https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&page_size=10&acc=SRR10971381&display=reads
https://www.metagenomics.wiki/tools/blast/blastn-output-format-6










Oh that is very, very interesting. I guess Wu or anyone else involved in the 2020 paper is not providing any answers. Here's little old me thinking that one should be able to replicate the results in published papers.
To me, publishing three different sequences should end it all right there.
Thanks for doing this work and sharing. It's big stuff.
I now figured out a less hacky way to show the positions where all sequences in an alignment don't have an identical value. It also works with three or more sequences, so for example this shows all the differences between the three versions of Wuhan-Hu-1:
curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.'{1,2,3}|mafft ->temp.aln
Rscript -e 'm=t(as.matrix(Biostrings::readDNAStringSet("temp.aln")));pick=which(rowSums(m!=m[,1])>0);write.table(`rownames<-`(m[pick,],pick),sep=";",quote=F,col.names=NA)'
Or the same in gawk:
awk 'NR>1{gsub("\t"," ")sub("\n","\t")gsub("\n","");print}' RS=\> temp.aln|awk -F\\t '{printf";%s",$1;l=length($2);for(i=1;i<=l;i++){x=substr($2,i,1);a[i][NR]=x;b[i][x]}}END{print"";for(i in b)if(length(b[i])>1){o=i;for(j=1;j<=NR;j++)o=o";"a[i][j];print o}}'